Dynamic Pattern Formation In Cells
The interior of any cell is far from homogeneous. What produces the patterns of protein distributions? Is there a map? Using the concept of Turing patterns, we are modelling protein systems that determine the site of cell division. We have identified a minimal set of biochemical reactions that via a reaction-diffusion mechanism create stable patterns that account for observed cell division. The model relates cell geometry to protein distributions. This is a collaboration with Chris Angstmann (UNSW). Additionally, we are collaborating with Iain Duggan and Liz Harry (University of Technology Sydney) to examine cell division in both archaea and bacteria.
Similar minimal models can be used to generate identifiers of axon selection in neurons. Neurons are comprised of a cell body with protrusions which receive and transmit electrical impulses. Dendrites are the primary receivers, of which there are many extending away from the cell body. Axons are the signal transmitters and there is typically only one per cell. Simple reaction diffusion systems yield a system that can identify the cells choice of a new axon (from it’s dendritic tree) after its existing axon is cut.
Nanomachines & Molecular Motors
Protein machines function as motors, but there is no satisfactory explanation as to how they transduce energy. We are part of an international group to try and build a protein based molecular motor from non-motor components (Heiner Linke, Lund University; Dek Woolfson, University of Bristol; Nancy Forde and Martin Zuckermann, Simon Fraser University; Beth Bromley, Durham University; Gerhard Blab, Utrecht University; and Till Böcking, UNSW). The aim is to test whether protein motors work by rectifying noise (Feynman-Smoluchowski Brownian ratchet). We have designed several candidate motors and we are currently building them using molecular biology and semi-synthetic approaches.
Molecular motors and machines are highly complex, multi-subunit proteins that use chemical energy to perform a multitude of critical, mechanical tasks in cells. The physical mechanisms by which motor proteins (such as myosin and kinesin) transduce chemical energy into mechanical work are still poorly understood. Traditionally, scientists have taken a “top down” approach to addressing this question by determining crystal structures of motor proteins, characterizing mutants and making single molecule measurements of performance.
The goal of our work is to take a “bottom up” approach and design an artificial motor based on non-motor protein components. In this way, we can test our understanding of motor protein operation by including components that have well characterized functional properties. To achieve this, we have set up an international collaborative team comprising our lab plus: Prof. Heiner Linke (Lund University, Sweden); A/Prof. Nancy Forde and Prof. Martin Zuckermann (Simon Fraser University, Canada) and Prof. Dek Woolfson (University of Bristol, UK). Our current design, the Tumbleweed, consists of a three-legged clocked walking protein that operates on a repetitive DNA track. Tumbleweed uses three discrete ligand-dependent DNA-binding domains (repressor proteins) to perform cyclical ligand-gated rectified diffusion along a synthetic DNA molecule.
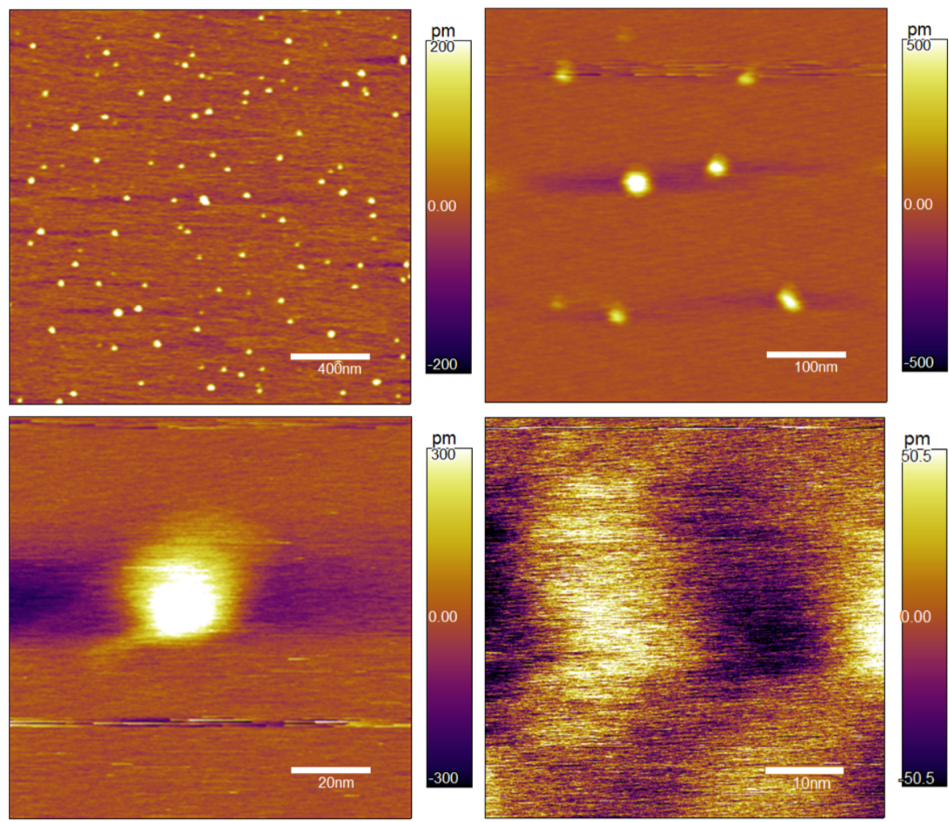
AFM image of tumbleweed attached to mica 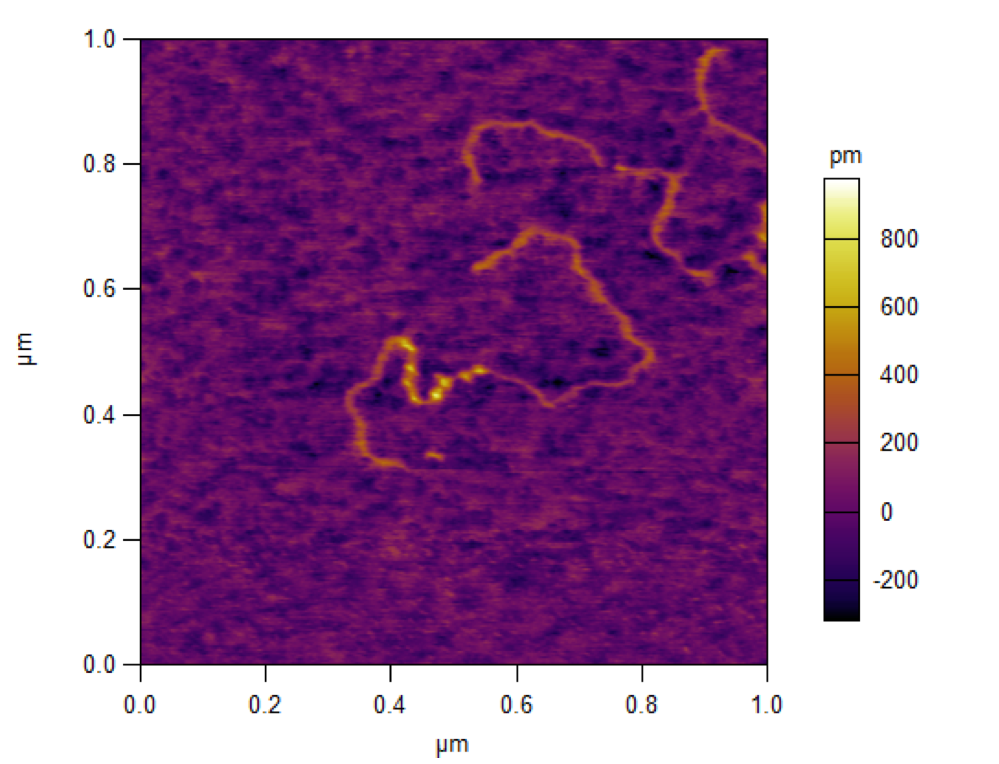
AFM image of tumbleweed attached to a DNA track
Non-trivial Quantum Effects In Biology
Quantum beats in the spectroscopy of light harvesting proteins
Proteins were assumed to function via classical physics with quantum effects playing trivial roles. In collaboration with Greg Scholes (University of Toronto) we have shown that certain algal light harvesting proteins show spectroscopic signatures of long-lived quantum coherence under near physiological conditions. Along with parallel discoveries, this has led to intense research activity aimed at determining the physical mechanism behind the observed long-lived oscillations in the 2D electronic spectra and whether quantum electronic coherence plays a role in photosynthetic light harvesting.
Cryptophyte light harvesting proteins
Cryptophytes are an unusual type of single-celled algae that have resulted from the endosymbiosis of a red algal cell inside a eukaryotic host. Like cyanobacteria and red algae, the cryptophytes have preserved a light harvesting system based on phycobiliproteins that are members of the globin fold superfamily. Unlike cyanobacteria and red algae, the cryptophyte phycobiliproteins are soluble and reside in the lumen on the thylakoid. We are using crystallography to unravel the mechanism by which these proteins trap light photons and transfer the energy to the membrane bound photosystem. We are collaborating with Greg Scholes (University of Toronto) and Jeff Davis (Swinburne University of Technology) whose groups are probing the light harvesting system via ultrafast laser spectroscopy. Our crystals of the light harvesting proteins diffract to ultra high resolution (<1Å).
Evolution of light harvesting proteins
The evolution of the cryptophyte light harvesting proteins has involved a radical rearrangement of the quaternary structure of the ancestral phycobilisome antenna complex from red algae and cyanobacteria. This rearrangement alters energy transfer within the light harvesting protein and between it and the membrane bound photosystems. We are tracking the evolution of light harvesting architectures within the cryptophytes in collaboration with Roger Hiller (Macquarie University), Beverley Green (University of British Columbia)and Kerstin Hoef-Emden(University of Cologne).Our aim is to characterise the structures of these complexes and determine the relationship between sequence, structure and spectroscopic properties.
4LMX-PE555-H.Andersenii 
4LMS-PC645-Chroomonas Sp. 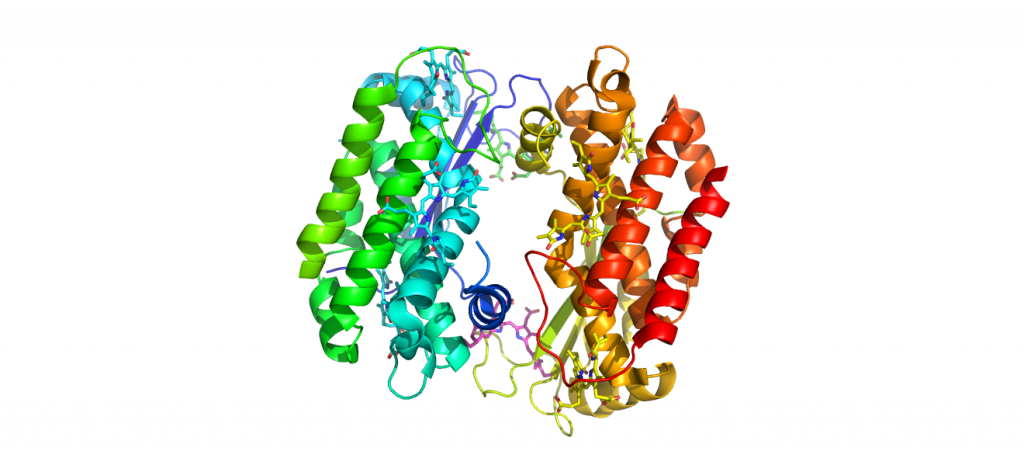
4LM6-PC612-H.Virescens 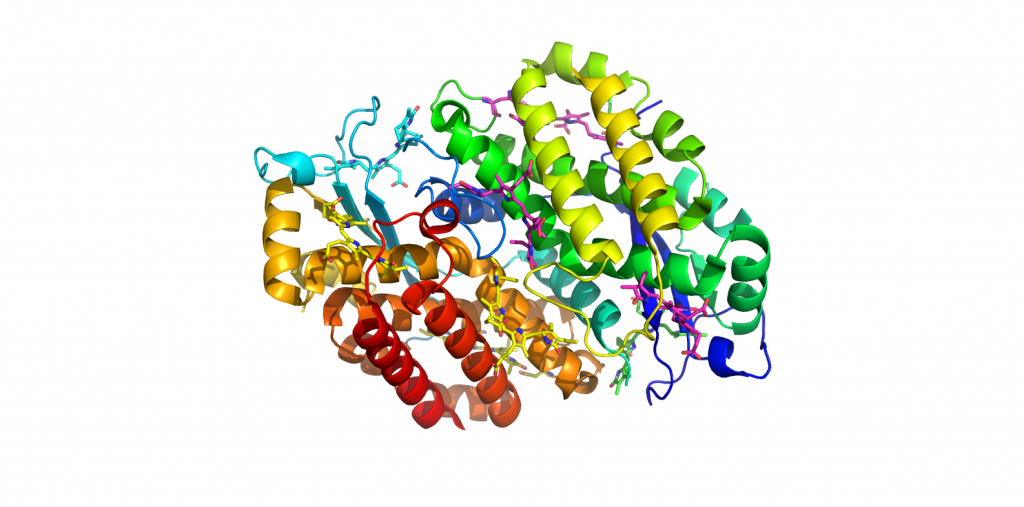
1XG0-PE545-Rhodomonas Sp.
Membrane Remodelling & Metamorphic Proteins
CLIC Proteins
CLIC proteins are unusual in that they exist in both globular and integral membrane states. The CLICs are highly conserved in vertebrates with homologues in most, if not all, metazoa. CLICs can form anion (chloride) channels in vitro and in vivo. Our goal is to gain a comprehensive understanding of the CLIC proteins in collaboration with Sam Breit (St. Vincent’s Centre for Applied Medical Research), Michele Mazzanti (University of Milan), Louise Brown (Macquarie University) and Stella Valenzuela (University of Technology Sydney). We have determined several crystal structures of CLIC proteins in the soluble form. In addition, we have discovered a dramatic structural change in CLIC1 that is stabilised by oxidation. Our current goal is to determine the structure of the integral membrane form of a CLIC protein as well as the structures of complexes with partner proteins.
Controlling membrane remodelling in mammalian cells
Cells are dynamic: they change shape, communicate with each other and import/export signalling molecules. These dynamic processes are controlled via the interaction of the cell membrane with the underlying actin cytoskeleton and they are important for health, for example, they are critical for proper immune cell function. The goal of this project is to unravel the control of membrane dynamics by defining the interactions between the cell membrane and the proteins: ezrin and RhoA.
The structure, activation and function of ezrin and RhoA are being pursued via a combination of structural biology (X-ray crystallography, small-angle X-ray scattering and neutron scattering) and single molecule fluorescence microscopy (collaboration with Dr Till Böcking, UNSW). By combining the results of these complementary approaches, we aim to understand how ezrin couples cortical actin filaments to membranes. We are exploring the role of other proteins in this system, including the CLIC proteins.
Metamorphic Proteins
It was believed that a protein sequence would define a single, well-defined three-dimensional structure. Using X-ray crystallography, we have discovered that the protein CLIC1 can adopt several well-defined structures. Along with the concurrent discovery of two other such proteins, this has led to the concept of metamorphic proteins. Currently we are focusing on understanding the structure, function and evolution of the CLIC protein family. In particular, we aim to determine the structure of the integral membrane state of the CLIC ion channel.
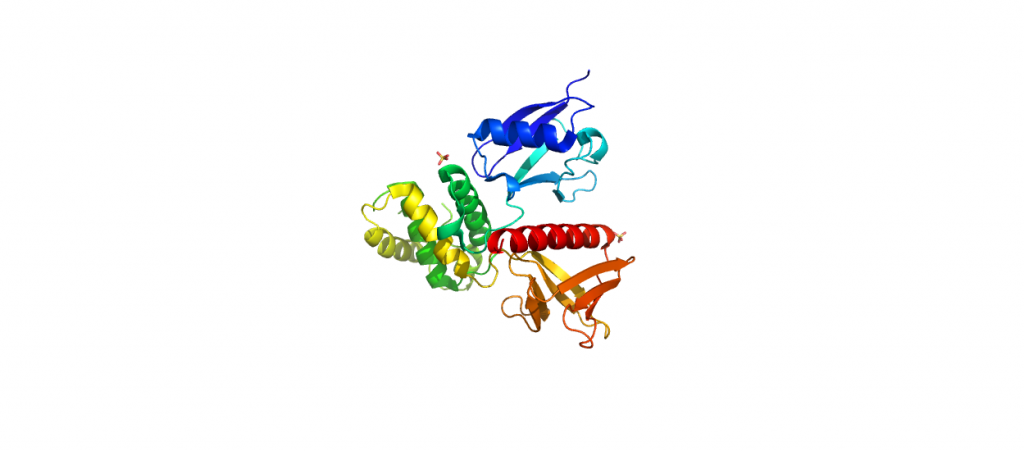
4RMA-Ezrin FERM 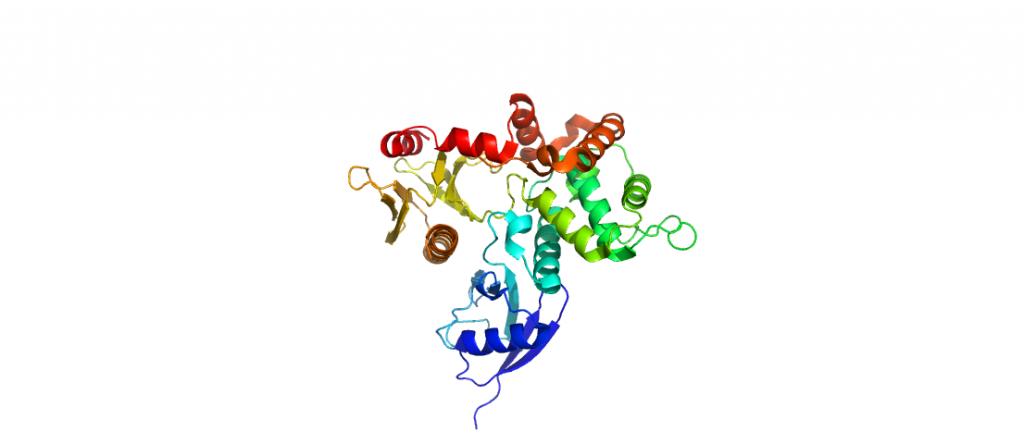
4RM9-Ezrin-C2221 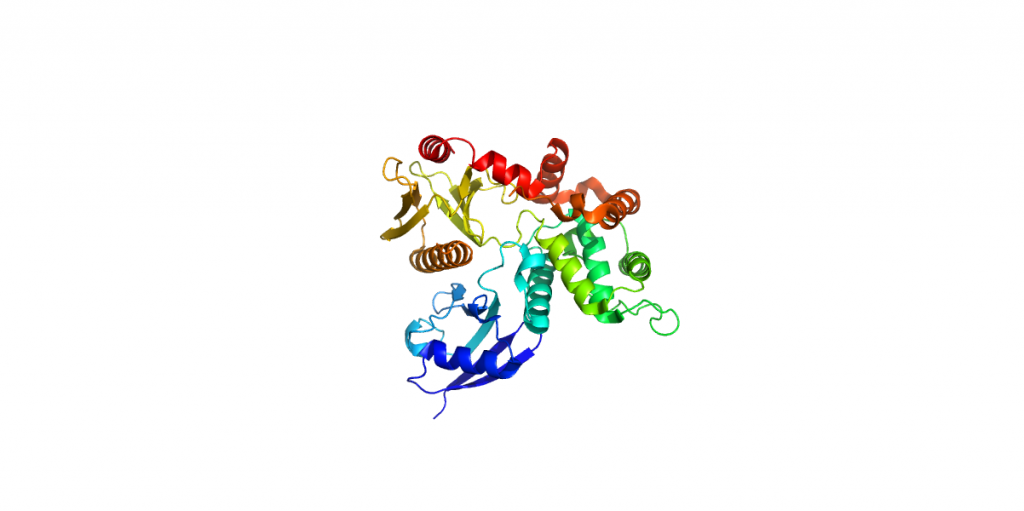
4RM8-Ezrin FERM 
4K0N-Clic1 
3OQS-Importan-alpha-bound-Clic4-NLS 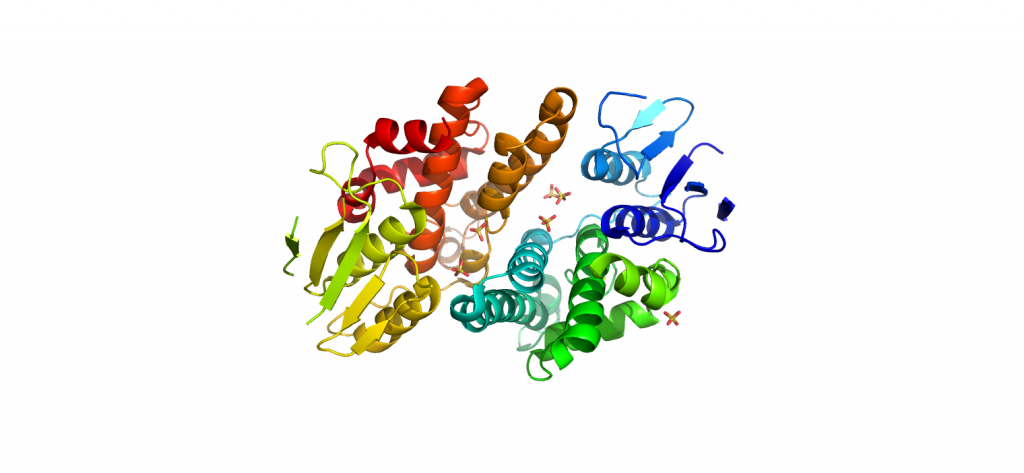
3KJY-Clic3-Reduced